
George Church (Harvard)
BIO DESIGN
Darwin on steroids: Design & Selection

DNA modification is fundamental to design in synthetic biology. For this week's assignment, you will learn to read DNA sequence files and see physical changes you will make to DNA molecules.
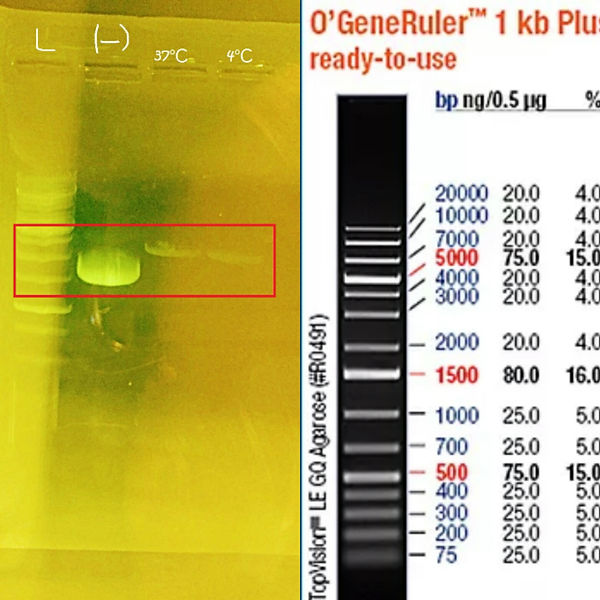
Lab Task: Verify restriction digest of a circular DNA plasmid by gel electrophoresis. Compare your experimental result with what you would predict from the plasmid map.
BRIEF PROTOCOL


PROCESS

RESULT
Título del sitio
Theory Task: Choose a DNA sequence that interests you from nature or a source like the iGEM part registry. Provide a detailed workflow for isolating or acquiring a molecule with this specific sequence and an assay that confirms part of its content.

Chinook aquareovirus, segment 11
Origin
1 gttttagaag atacaggagc agcaatggca tacaaaccca tcatcgatgt aaccgaacta
61 gcccaacaac ttataaacac taaacttttc tcctccgcca aagatgtctc atccatccgt
121 agagagctgt cgcgtcacgc tgcagaccca tatagtcatg agcacactgt tggagactcc
181 ccccctccca ctccctacct caatcaggtc ccaagcagtt cgtataggct ttgtgctgtg
241 ccggtcgtgt cacattccag tggaggtcat tccgcaaccg tgtctgtgcc agctcgagtc
301 attgttgatg ttactgcagc caccctcgtc ctcacgttcc catcctacga cttccacgtc
361 caccaaccac tagtctcagg cggcaagctg tttctgtcca tcgacaaggg caattggcat
421 gacctgtcta ctgggtgctt caacctcgac gccttcaaca tgctgagaga taccgccact
481 ggtaagaatt ccaccaccgt cagcgttaaa tggtacggcg gcactggacc atcccgtgga
541 gatcttcgct caacaggctc tcgcgcccgt attcatgtga ccgaccgtcg catcatgttc
601 gagctgcctg gagtcgactc ggctggcacc tatcccgacc taaccgtgtg gggctcgttc
661 gcagtattcg agtaggcgcg tgtgtccttg ctggcgtggc cgcgtaaacc ctgtatctat
721 cattcatc
WORKFLOW TO ISOLATE THIS SEQUENCE
Título del sitio

In this lab task we learned about the enzyme restriction function, DNA plasmid, gel electrophoresis and how it works. Also, we compared our results of this practice with the plasmid map.
It is important to read about the materials that will be used for the development of the practice and ask yourself the following questions:
-
What is a restriction enzymes?
A restriction enzyme or restriction endonuclease is an enzyme that cleaves DNA into fragments at or near specific recognition sites within the molecule known as restriction sites. After making these cuts, we will run our samples in an electrophoresis gel and we will see the differences of size that exist.
-
What is a gel?
As the name suggests, gel electrophoresis involves a gel: Gels for DNA separation are often made out of agarose, this is a polysaccharide polymer material, generally extracted from seaweed, which comes as dry, powdered flakes.
-
How to prepare the gel?
For this practice we used 1% gel. This consisted in mix 1g of the gel in 100Ml of buffer TAE.
Buffers are used to provide ions that carry a current and to maintain the pH at a relatively constant value.
Most agarose gels are made with between 0.7% (good separation or resolution of large 5-10kb DNA fragments) and 2% (good resolution for small 0.2-1kb fragments) agarose dissolved in electrophoresis buffer. Low percentage gels are very weak and may break when you try to lift them. High percentage gels are often brittle and do not evenly set. 1% gels are common for many applications
-
What is a gel electrophoresis?
Gel electrophoresis is a method for separation and analysis of macromolecules (DNA, RNA and proteins) and their fragments, based on their size and charge. It is used in clinical chemistry, biochemistry and molecular biology to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments or to separate proteins by charge.
-
How does it work?
Before the DNA samples are added, the gel must be placed in a gel box. One end of the box is hooked to a positive electrode, while the other end is hooked to a negative electrode. The main body of the box, where the gel is placed, is filled with a salt-containing buffer solution that can conduct current, the buffer fills the gel box to a level where it just barely covers the gel.
The end of the gel with the wells is positioned towards the negative electrode. The end without wells is positioned towards the positive electrode.
Smaller molecules migrate through the gel more quickly and therefore travel further than larger fragments that migrate more slowly and therefore will travel a shorter distance. As a result the molecules are separated by size.
A dye is added to the sample of DNA prior to electrophoresis to increase the viscosity of the sample which will prevent it from floating out of the wells and so that the migration of the sample through the gel can be seen.
Do not forget to put the dye to your gel before starting to load the samples. The use of dyes enables the DNA on the gel to be seen after they have been separated.
To visualise the DNA, it is important that the gel is with a fluorescent dyed that binds to the DNA, and is placed on an ultraviolet transilluminator which will show up the stained DNA as the bright bands.
-
How do DNA fragments move through the gel?
Once the gel is in the box, each of the DNA samples we want to examine (for instance, each restriction-digested plasmid) is carefully transferred into one of the wells. One well is reserved for a DNA ladder, a standard reference that contains DNA fragments of known lengths.
Next, the power to the gel box is turned on, and current begins to flow through the gel. The DNA molecules have a negative charge because of the phosphate groups in their sugar-phosphate backbone, so they start moving through the matrix of the gel towards the positive pole. When the power is turned on and current is passing through the gel, the gel is said to be running.
As the gel runs, shorter pieces of DNA will travel through the pores of the gel matrix faster than longer ones. After the gel has run for awhile, the shortest pieces of DNA will be close to the positive end of the gel, while the longest pieces of DNA will remain near the wells
Once the fragments have been separated, we can examine the gel and see what sizes of bands are found on it. When a gel is stained with a DNA-binding dye and placed under UV light, the DNA fragments will glow, allowing us to see the DNA present at different locations along the length of the gel
By comparing the bands in a sample to the DNA ladder, we can determine their approximate sizes
-
When is gel electrophoresis used to separate DNA fragments?
Gel electrophoresis can be used for a range of purposes, for example:
-
To get a DNA fingerprint for forensic purposes
-
To get a DNA fingerprint for paternity testing
-
To get a DNA fingerprint so that you can look for evolutionary relationships among organisms
-
To check a PCR reaction.
-
To test for genes associated with a particular disease.







